Using ColabFold to Predict Protein Structures

If I know the sequence of amino acids, how do I figure out the three-dimensional structure and as a result the function of a protein? The concept of a protein folding problem has been around since 1960, with the appearance of the first atomic-resolution protein structures.
The release of AlphaFold 2 three years ago marked a massive step in solving the protein folding problem, determining the structures of around 200 million proteins. We’re going to examine AlphaFold 2, how it was able to achieve such spectacular results, and a particular implementation of AlphaFold 2 — ColabFold — that I used to predict the structures of proteins. I used ColabFold mainly because it was less memory intensive as compared to AlphaFold 2, which required over 3 TB of storage to run. Besides being less memory intensive, ColabFold is virtually the same thing as AlphaFold 2, except that it combines AlphaFold 2 with the fast homology search of MMseqs2 (Many-against-Many sequence searching). MMseq2 is a software that performs fast and deep clustering and searching of large datasets. Similarity clustering reduces the redundancy of sequence databases, improving speed and sensitivity of iterative searches. MMseqs can cluster large databases down to around 30% sequence identity at hundreds of times the speed of BLASTclust and much deeper than CD-HIT and USEARCH. In combining AlphaFold 2 and MMseq2, researchers can leverage both sequence similarity information and structural predictions to gain insights into the functions and properties of proteins.
Why Predict Protein Structure?
Why does this problem even matter in the first place? Well, here are three big reasons why this problem is worth solving.
Structure-Based Drug Discovery
One of the biggest implications of predicting the structure of proteins is for structure-based drug discovery, where using the structure of the protein, candidate drugs are picked based on if they can bind to the target with high affinity and selectivity. Affinity refers to the strength of attraction between the drug and its receptor, with a high affinity indicating a lower dose requirement. Drugs that are highly selective are also favorable because it means that they mainly affect a single organ or system, reducing unwanted side-effects that usually occur when a drug affects other parts of the body. This is especially crucial for drugs treating chronic conditions where long-term use is necessary.
Knowing a proteins structure is especially important for binding site identification — the first step in structure-based drug discovery. Here, scientists look for concave surfaces (surfaces that are curved inwards) on a protein that can fit drug-sized molecules. These areas need to have special surfaces (hydrophobic, hydrophilic, etc) that make them attractive for molecules to bind to, ensuring effective that the drug will stick. If we do not know the structure of a protein, how do we expect scientists to find these areas that a drug can potentially bind to?
Engineering Proteins
Related also to drug discovery, but if we can predict the structure of a protein in a short timeframe — days, rather than months or years — we could work backwards and first design a desired structure for a protein, specifically an active site, and then try to fine-tune a protein sequence to adopt this structure. This means that instead of adopting our drugs to fit the structure of a protein, we could design proteins to fit the structure of our therapies.
Understanding Protein Function
Protein function is dependent on its structure. If we learn more about a proteins structure, we can gain a better understanding of its function, and ultimately help us answer the question: “What is life?”
AlphaFold 2 (AF2)
A few hours before the CASP14 (14th Critical Assessment of Structure Prediction) meeting, the latest biannual structure prediction experiment where participants build models of proteins given their amino acid sequences, this image went viral on twitter.

As you can see, one group, 427, absolutely destroyed the competition: that group was AlphaFold 2. Clearly AlphaFold 2 was ahead of the game, truly being more advanced in terms of how it predicted protein structure.
Just How Good was AlphaFold 2?
To provide a little more context on how good AlphaFold 2 was, here are some other statistics that clearly showcase AlphaFold 2’s incredible performance.
Using a value called Root-mean-square deviation (RMSD) of atomic positions, which is a way to measure the the average distance between the atoms of superimposed molecules. RMSD allows us to know how different or similar two molecules are, giving us a picture of if the predicted protein structure is a indeed an accurate prediction. In proteins, we focus on certain atoms (Cα atoms) and measure the RMSD of their positions after aligning the molecules in the best possible way. For reference, a lower RMSD represents a better predicted structure, and most experimental structures have a resolution around 2.5 Å
For AlphaFold 2, about a third (36%) their submitted targets were predicted with RMSD under 2 Å, and 86% were under 5 Å, with a total mean of 3.8 Å.

Using a value called GDT_TS, AlphaFold 2 was also compared to the results of all previous CASP competitions. For reference, a GDT_TS of 100 represents perfect results and 0 is a meaningless prediction. A GDT_TS around 60% represents a “correct fold”, meaning that we have an idea of how the protein folds globally; and over 80% we start seeing side chains that closely resemble the model. As you can see, AlphaFold 2 achieves this objective for all but a small percentage of the tasks.

In case you aren’t already convinced, I wanted to show a few examples of some of the predictions made by AlphaFold 2.
Here is the prediction of the ORF8 protein, a viral protein involved in the interaction between SARS-CoV-2 and the immune response (labeled as T1064).

What impressive is how AlphaFold 2 predicted the loops. Loops are types of secondary protein structures that connect α-helices and β-strands, the two most common types of secondary structures in a protein. Both structures are held in shape by hydrogen bonds, which form between the carbonyl O of one amino acid and the amino H of another.

Loops usually cause a change in direction of the polypeptide chain, allowing the protein to fold back on itself to create a more compact structure. Loop regions are characterized by a lack of secondary structure, meaning that there is not a scaffold of hydrogen bonds that maintains the structure together, as opposed to α-helices and β-sheets. Thus, the structure of loops is far more flexible and generally considered harder to predict, showing how impressive AlphaFold 2’s performance was.
Another example is the target T1046s1. In this case, AlphaFold 2 is virtually indistinguishable from the crystal structure with an impressive total RMSD of 0.48 Å!

After three decades of competitions, the assessors declared that AlphaFold 2 had succeeded in solving a challenge open for 50 years: to develop a method that can accurately predict a protein structure from its sequence. While there are caveats and edge cases, the magnitude of the breakthrough as well as its potential impact are undeniable.
How it Works
So how did AlphaFold 2 achieve these incredible results?
The first step is that AlphaFold 2 system uses the input amino acid sequence to query several databases of protein sequences, and constructs a multiple sequence alignment (MSA). AlphaFold 2 comes equipped with a “preprocessing pipeline”, which is just a bash script that calls some other code. The pipeline runs a number of programs for querying databases and, using the input sequence, generates a MSA and a list of templates.
Templates are proteins that may have a similar structure to the input. These templates are used to construct an initial representation of the protein structure, which it calls the pair representation. This is, in essence, a model of which amino acids are likely to be in contact with each other.
A MSA is the the result of aligning three or more biological sequences, generally either a protein, DNA, or RNA. In AlphaFold’s case, it finds multiple similar proteins and aligns their sequence together with the target protein’s sequence.

To find similar sequences, the sequence of the protein whose structure we intend to predict is compared across a large database (normally something such as UniRef, although in later years it has been common to enrich these alignments with sequences derived from metagenomics). Then, JackHMMER, HHBLITs, or PSI BLAST are used within the MSA to maximize scores and correctness of the alignments.
What the MSA is ultimately looking for are conserved regions and co-evolutionary patterns. Conserved regions in a protein sequence refer to segments of the amino acid sequence that remain relatively unchanged across different species or variants over evolutionary time. Proteins mutate and evolve, but their structures tend to remain similar despite the changes. Conserved regions usually occur on a small scale, where pieces of the protein (for example, the active center of an enzyme) remain mostly unchanged while their surroundings evolve. By aligning the sequences of related proteins, researchers can pinpoint positions where the amino acids remain unchanged, indicating conservation. These conserved regions indicate that certain positions in the amino acid sequence are crucial for the structural and functional integrity of the protein, serving as a guide to construct the structure of the target protein.
To illustrate conservation, let us take the structures of four different myoglobin proteins, corresponding to different organisms. You can see that they all look pretty much the same, but if you were to look at the sequences, you would find enormous differences. The protein on the bottom right, for example, only has around 25% amino acids in common with the protein on the top left.

The next step is to take the MSA and the templates, and pass them through a transformer. AlphaFold’s transformed is called the Evoformer, which has the task of essentially getting all of the information out of the MSA and the templates. In the Evoformer, information flows back and forth throughout the network.
Before AlphaFold 2, most deep learning models would take a MSA and output some inference about geometric proximity of elements in a sequence. Geometric information was therefore a product of the network. In the Evoformer, the pair representations is a both a product and an intermediate layer.
At every cycle, the model leverages the current structural hypothesis to improve the assessment of the MSA, which in turns leads to a new structural hypothesis, and so on, and so on. Both representations, sequence and structure, exchange information until the network reaches a solid inference and can not be tangibly improved anymore.
For example, suppose that you look at the MSA and notice a correlation between a pair of amino acids: A and B. You hypothesize that A and B are close, and translate this assumption into your model of the structure. Subsequently, you examine said model and hypothesize that, since A and B are close, there is a good chance that C and D should be close. This hypothesize can then be confirmed by looking back at the MSA and searching for correlations between C and D. By repeating this several times, you can build a pretty good understanding of the structure.
Lets dive into a more technical explanation of the Evoformer because I think that it is so interesting.
The first step in the Evoformer is to define embeddings for the MSA and templates. Essentially, an embedding is a technique that allows the transformation of a discrete variable (MSA) to a continuous/embedded space so that the network can be properly trained and the Evoformer can work its magic. MSA’s are a discrete variable because they are ultimately sequences of symbols (A, G, C, and T) from a finite alphabet. On the other hand, neural networks are intrinsically continuous devices that rely on differentiation to learn from their training set.
The way that this embedding is done is by defining a layer of neurons that receives the discrete input and outputs some continuous vector. In AlphaFold 2, the embeddings are vanilla dense neural networks.
The next step of the Evoformer is to identify which parts of the input are the most important to pay attention to. Transformers all have an attention mechanism, which identifies which parts of the input are more important for the objective of the neural network.
Lets take an example, imagine that you are trying to train a neural network to produce image captions. One possible approach is to train the network to process the whole image, which could be around 250k pixels in a 512×512 picture. This may work with smaller images, but there are some reasons why it is not the best idea for larger pictures. First of all, because this is not what we do: when you look into a picture, you do not see it “as a whole”. Instead, even if you do not realize it, we focus on key elements of the image: a child, a dog, a frisbee. Luckily, we can train a cleverly-designed neural network layer to perform this exact task. And, empirically, it seems to improve the performance by a lot.

Bringing this back to proteins attention reveals which parts of the sequence are important for the current part of the translation. The only reason why transformers are not widely adopted is because the construction of the attention matrix leads to a quadratic memory cost, which may not be economical for a lot of researchers.
The Evoformer has two transformers with one clear communication between the two. One transformer is focused on the MSA while the other is focused on pair representations. They also regularly exchange information in order to refine the data that they should focus on.
For the one focused on the MSA. The network first computes attention in the horizontal direction, allowing the network to identify which pairs of amino acids are more related; and then in the vertical direction, determining which sequences are more informative. This reduces what would be otherwise an impossible computational cost.
For the one focused on pair representations, attention is arranged in terms of triangles of residues.

The Evoformer process is organized in blocks that are repeated iteratively until a specified number of cycles (48 blocks in the published model).
The final step is to actually generate the model. Here, AlphaFold 2 takes the refined MSA and pair representation and leverages them to construct a three-dimensional model of the structure. Unlike the previous state-of-the-art models, AlphaFold 2 does not use any optimization algorithm. Instead, it generates a static, final structure, in a single step. The end result is a long list of Cartesian coordinates representing the position of each atom of the protein, including side chains.
The way AlphaFold 2 does this is by modeling every amino acid as a triangle. These triangles float around in space, and are moved by the network to form the structure. These transformations are parameterized as “affine matrices,” which are a mathematical way to represent translations and rotations in a single 4×4 matrix. The key is a system called Invariant Point Attention (IPA), devised specifically for working with three-dimensional structures.
After generating a final structure, the MSA, pair representation, and predicted structure will all be passed back to the beginning of the Evoformer blocks in order to refine the prediction.
Summary
I realize I just threw a lot of information at you so let me take a step back and quickly summarize. The main steps to AlphaFold 2 are:
- Finds similar sequences to the target protein and construct a MSA
- Use templates to construct a pair representation
- Pass MSA and pair representation into the Evoformer
- Generate structure using refined MSA and pair representation
- Pass MSA, pair representation, and generated strucutre back to Evoformer and repeat steps 3–5

Limitations
Although it is incredibly advanced, AlphaFold 2 is not a perfect model, here are some existing limitations that must be acknowledged:
- AlphaFold depends on a multiple sequence alignment as input, and it remains to be seen if it can tackle problems where the inputs are shallow or not very informative, as happens with designed proteins or antibody sequences. The DeepMind team suggests that “accuracy drops substantially when the mean alignment depth is less than around 30 sequences.”
- Hasn’t solved membrane proteins.
- While AlphaFold 2 has solved one facet of the problem — predicting structure from sequence, they didn’t solve the more fundamental question of **how the protein folds. If they understand this, they can as a result understand how proteins misfold, which is the biggest reason Alzheimer’s and Parkinson’s exist. Only until we understand how proteins fold can we being to dream about curing those diseases.
Results
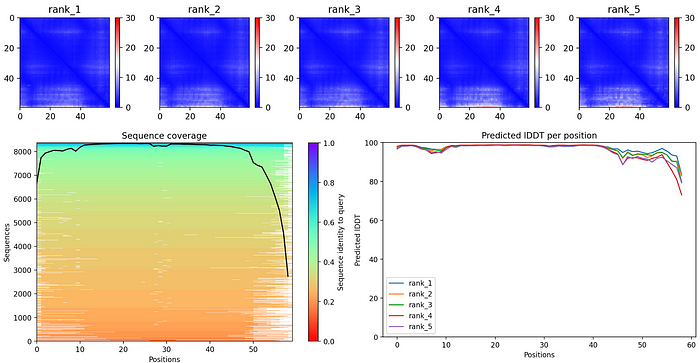
I tried predicting some protein structures myself using ColabFold. I used UniProt to find my protein sequences and played around with all the different options that I could customize. I definitely recommend making sure to show the side and main chains but beyond that I left most of the parameters untouched in the end.
The results were overall very good and I got a lot of accurate protein structures. If you want to check it out and try it yourself, here is the Github and Google Colab. Here is an example:
Query sequence: PIAQIHILEGRSDEQKETLIREVSEAISRSLDAPLTSVRVIITEMAKGHFGIGGELASK

Before you go
If you liked this article, have any questions, or want to provide any feedback, please email me (natank929@gmail.com) or check out my LinkedIn to connect further.
Sources
I wanted to again highlight some of the the best articles that I read to help inform my research:
- https://www.nature.com/articles/s41586-021-03819-2
- https://www.nature.com/articles/s41592-022-01488-1#:~:text=ColabFold enables researchers to upload,from the HH-suite8.
- https://www.blopig.com/blog/2021/07/alphafold-2-is-here-whats-behind-the-structure-prediction-miracle/
- https://www.blopig.com/blog/2020/12/casp14-what-google-deepminds-alphafold-2-really-achieved-and-what-it-means-for-protein-folding-biology-and-bioinformatics/
